cKBET: assessing goodness of batch effect correction for single-cell RNA-seq
Por um escritor misterioso
Last updated 13 junho 2024

lt;p>Single-cell RNA sequencing reveals the gene structure and gene expression status of a single cell, which can reflect the heterogeneity between cells. However, batch effects caused by non-biological factors may hinder data integration and downstream analysis. Although the batch effect can be evaluated by visualizing the data, which actually is subjective and inaccurate. In this work, we propose a quantitative method cKBET, which considers the batch and cell type information simultaneously. The cKBET method accesses batch effects by comparing the global and local fraction of cells of different batches in different cell types. We verify the performance of our cKBET method on simulated and real biological data sets. The experimental results show that our cKBET method is superior to existing methods in most cases. In general, our cKBET method can detect batch effect with either balanced or unbalanced cell types, and thus evaluate batch correction methods.</p>
A test metric for assessing single-cell RNA-seq batch correction

Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors
How to Batch Correct Single Cell. Comparing batch correction methods for…, by Nikolay Oskolkov
Fast, sensitive and accurate integration of single-cell data with Harmony
A benchmark of batch-effect correction methods for single-cell RNA sequencing data, Genome Biology

A benchmark of batch-effect correction methods for single-cell RNA sequencing data, Genome Biology

cKBET: assessing goodness of batch effect correction for single-cell RNA-seq

Removal of Batch Effects from Single-cell RNA-Seq
How to Batch Correct Single Cell. Comparing batch correction methods for…, by Nikolay Oskolkov

PDF] Projected t-SNE for batch correction

cKBET: assessing goodness of batch effect correction for single-cell RNA-seq
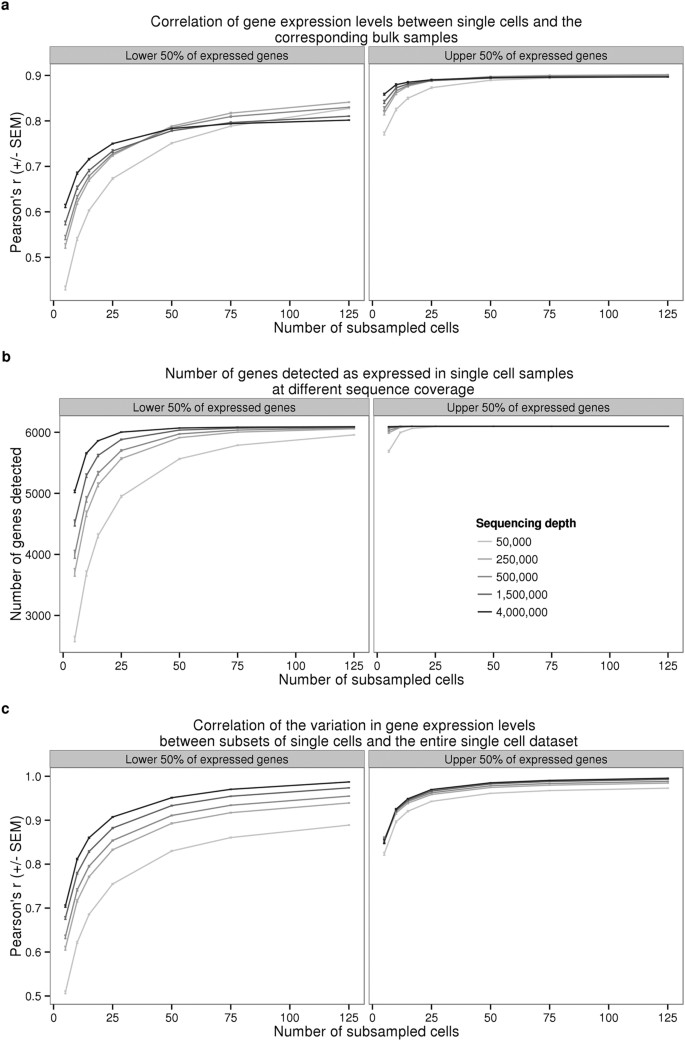
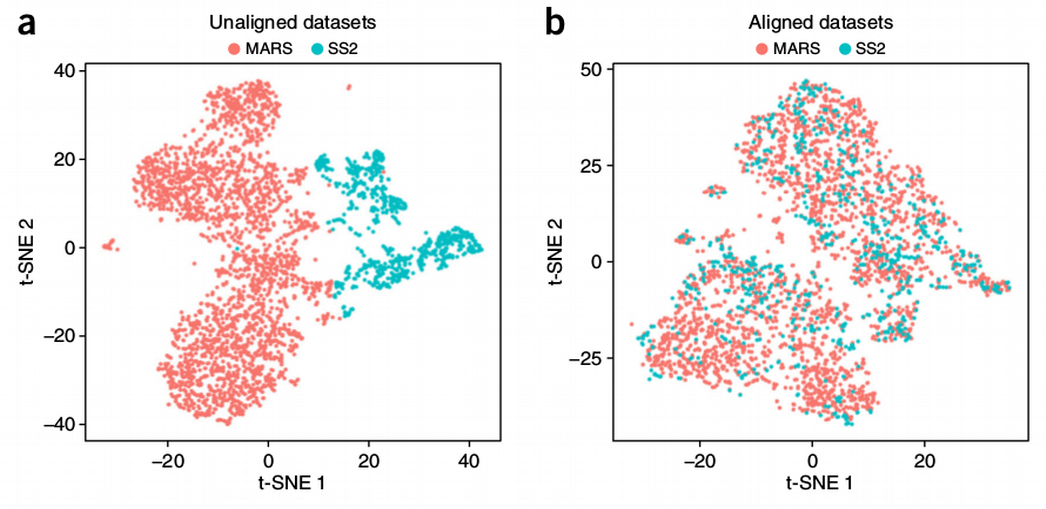
Batch effects and the effective design of single-cell gene expression studies
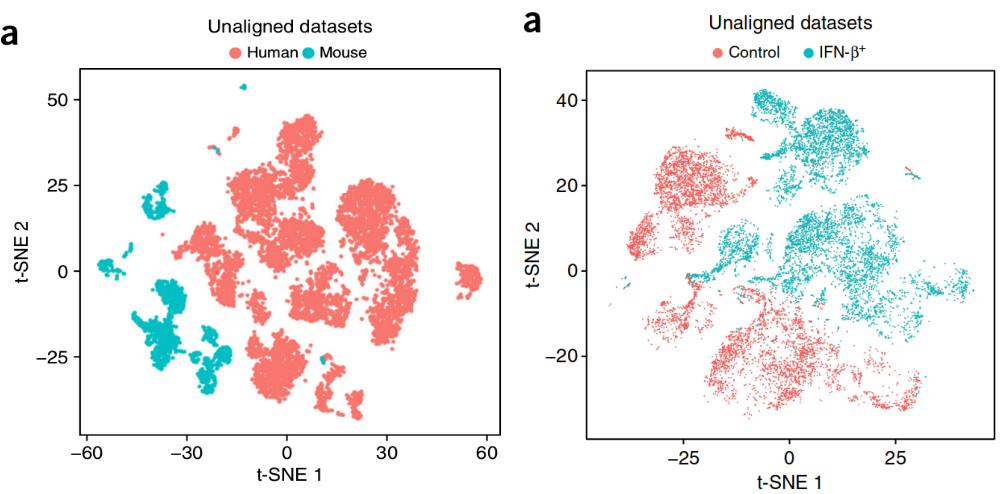
How to Batch Correct Single Cell. Comparing batch correction methods for…, by Nikolay Oskolkov
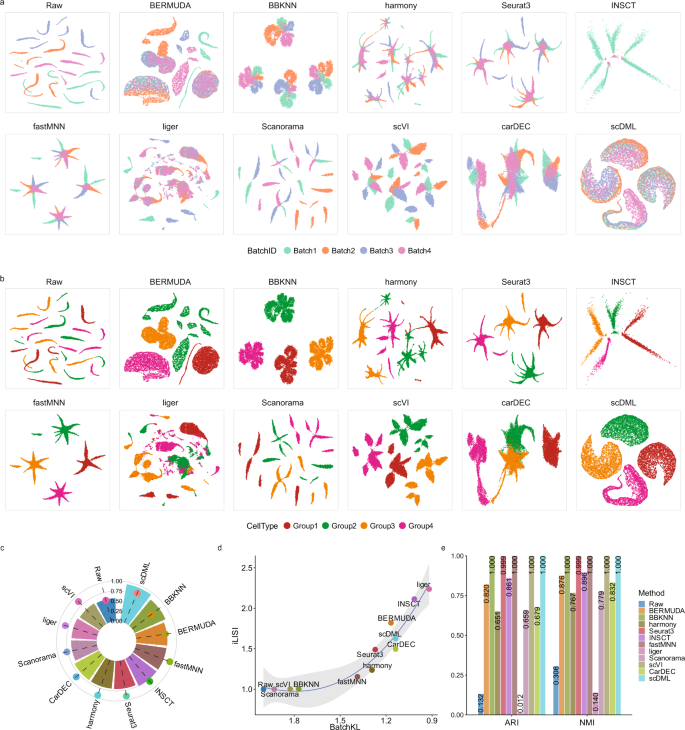
Batch alignment of single-cell transcriptomics data using deep metric learning
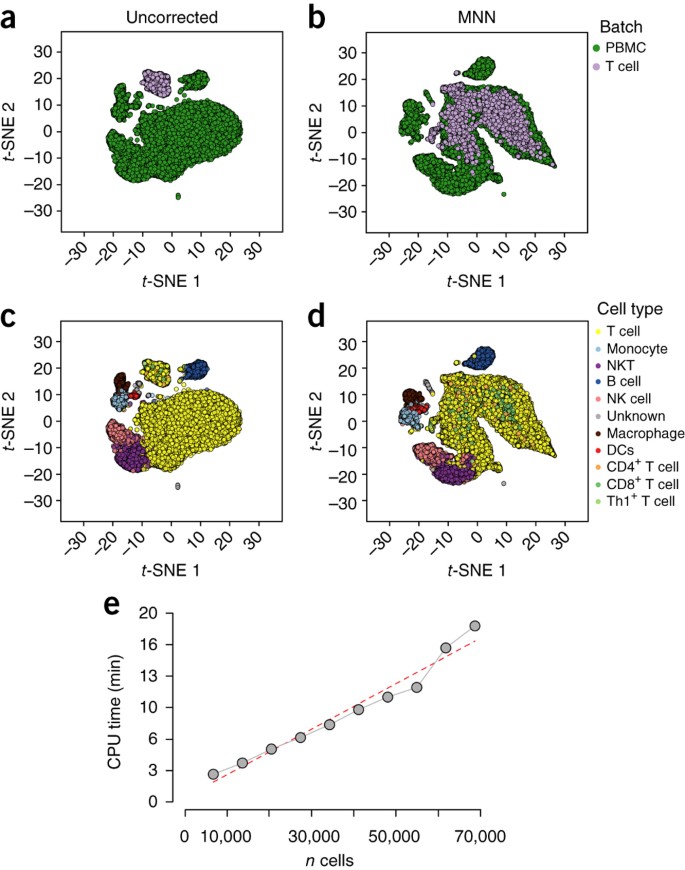
Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors
Recomendado para você
-
 A Plataforma de JOGOS DE CASSINO ONLINE Mais TOP e Mais Confiável!13 junho 2024
A Plataforma de JOGOS DE CASSINO ONLINE Mais TOP e Mais Confiável!13 junho 2024 -
 CKBet - casa de apostas com apostas online13 junho 2024
CKBet - casa de apostas com apostas online13 junho 2024 -
 Ckbet10113 junho 2024
Ckbet10113 junho 2024 -
 Odd_Limit_6518 (u/Odd_Limit_6518) - Reddit13 junho 2024
Odd_Limit_6518 (u/Odd_Limit_6518) - Reddit13 junho 2024 -
 Ckbet.com em 2023 Jogos online, Cassino, Jogos13 junho 2024
Ckbet.com em 2023 Jogos online, Cassino, Jogos13 junho 2024 -
 Browse thousands of 168jogo Com Sacar Ckbet Com95429 images for design inspiration13 junho 2024
Browse thousands of 168jogo Com Sacar Ckbet Com95429 images for design inspiration13 junho 2024 -
Download Ckbet on PC (Emulator) - LDPlayer13 junho 2024
-
CKbet - Comissão de Membros CKBET Seja um agente da CKBET, convide seus amigos para fazerem parte da sua equipe e obtenha sua comissão sem nem precisar apostar ou jogar nada! Contanto13 junho 2024
-
 ckbet13 junho 2024
ckbet13 junho 2024 -
 HOME Ckbet13 junho 2024
HOME Ckbet13 junho 2024
você pode gostar
-
 Heavy Club Chess Set13 junho 2024
Heavy Club Chess Set13 junho 2024 -
Tudo que você precisa saber sobre a Pokémon GO Tour 2022: Johto13 junho 2024
-
 Jonathan's mixtapes, Stranger Things Wiki13 junho 2024
Jonathan's mixtapes, Stranger Things Wiki13 junho 2024 -
_still-working-on-2023roblox-audio-music-boombox-songids-robloxid-song-robloxcodes-games-preview-hqdefault.jpg) Still working on 2023🤭!#roblox #audio #music #boombox #songids13 junho 2024
Still working on 2023🤭!#roblox #audio #music #boombox #songids13 junho 2024 -
 melhorcasa de aposta13 junho 2024
melhorcasa de aposta13 junho 2024 -
 Carrinho Hot Wheels 12 Peças - Diversos Modelos - C4982 em13 junho 2024
Carrinho Hot Wheels 12 Peças - Diversos Modelos - C4982 em13 junho 2024 -
 Kurumi Tokisaki Light Novel Date Ver Date A Live Figure13 junho 2024
Kurumi Tokisaki Light Novel Date Ver Date A Live Figure13 junho 2024 -
 A Quick Look at the Summer 2023 Anime Season : r/anime13 junho 2024
A Quick Look at the Summer 2023 Anime Season : r/anime13 junho 2024 -
 Nomad's blog: The Last Guardian - Thoughts & Comparisons13 junho 2024
Nomad's blog: The Last Guardian - Thoughts & Comparisons13 junho 2024 -
 De onde é essa tirinha do Zangado? É do livro? : gamesEcultura13 junho 2024
De onde é essa tirinha do Zangado? É do livro? : gamesEcultura13 junho 2024

