Accurate proteome-wide missense variant effect prediction with AlphaMissense
Por um escritor misterioso
Last updated 03 julho 2024


Evaluating gMVP and published methods using cancer somatic mutation

DeepMind launches AI tool Alpha Missense to predict harmful genetic mutations in humans - Hayo AI tools

Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated?

Combining AlphScore with established missense prediction scores
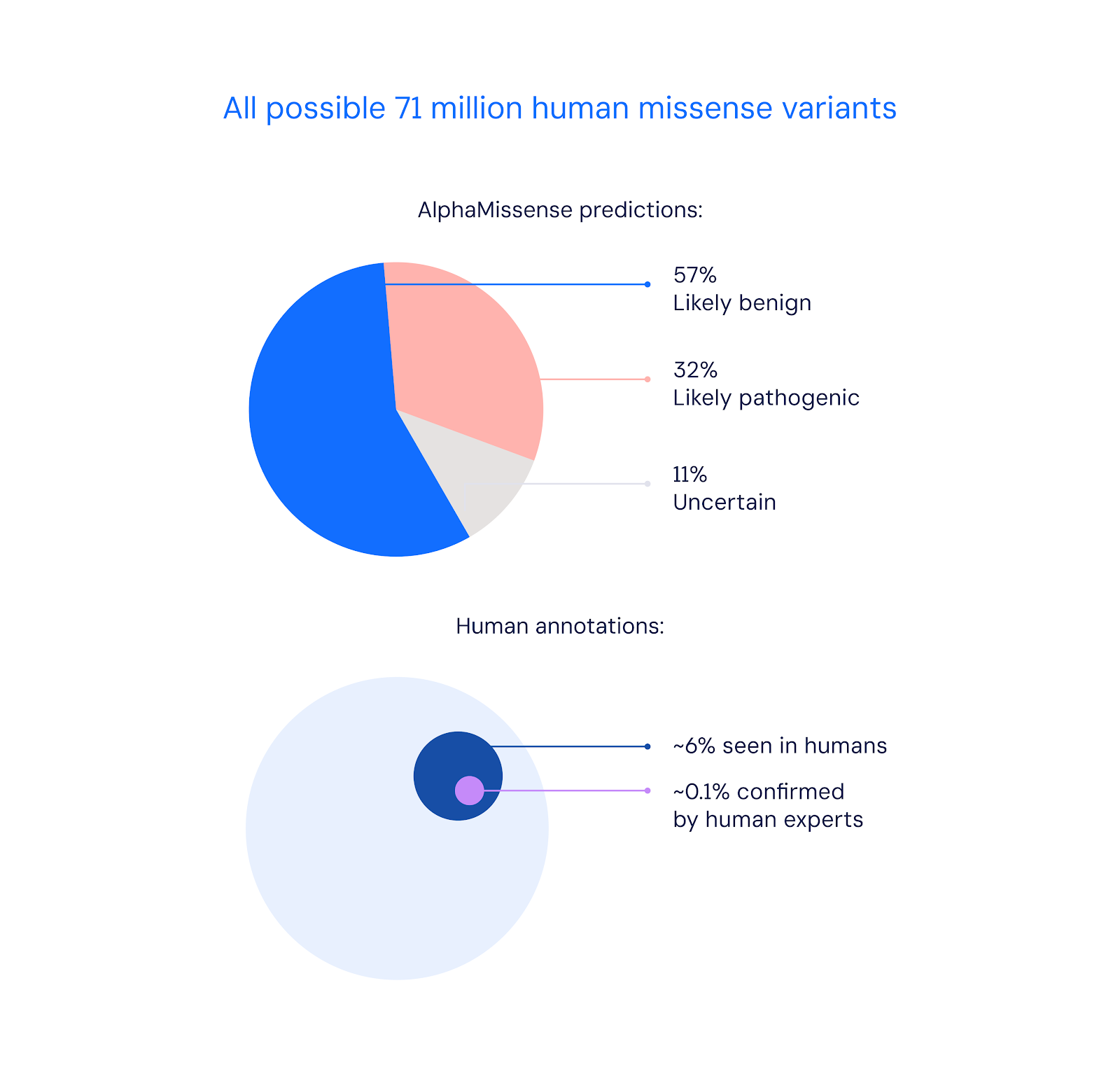
Predicting variant pathogenicity with AlphaMissense
Meet AlphaMissense: Google's New AI Tool that Can Identify Protein Mutations Likely to Cause Disease
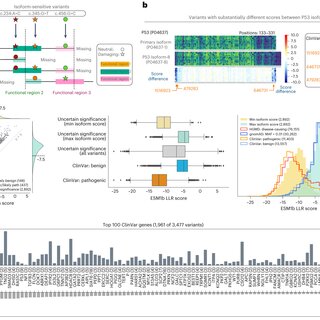
ESM1b predictions in clinically relevant genes depend on the isoform

Google DeepMind Says Its New Artificial Intelligence Tool Can Predict Which Genetic Variants Are Likely to Cause Disease
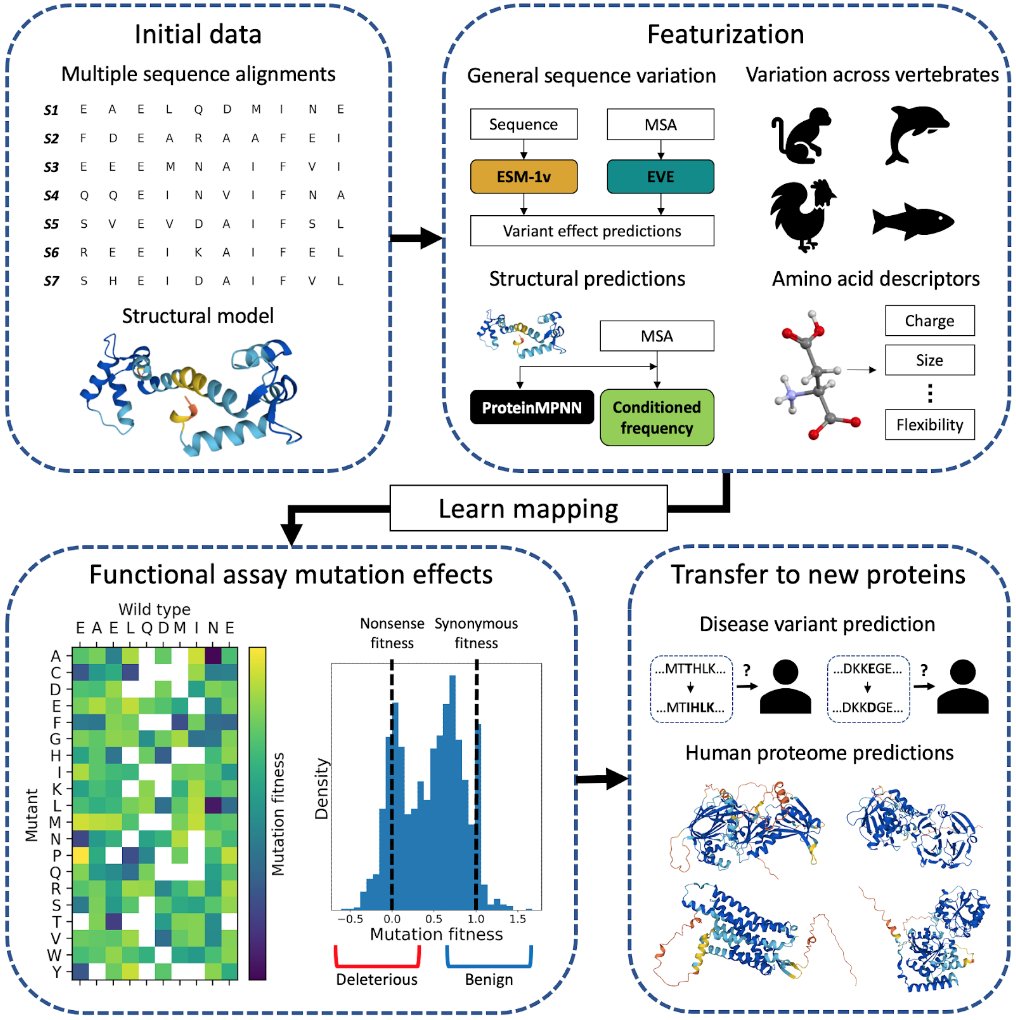
Yun S. Song on X: Predicting the effects of missense variants is a central problem in human genome interpretation. We are thrilled to share our preprint on using cross-protein transfer (CPT) learning
Recomendado para você
-
 Where Is Thomas Mitchell Overton Now? Learn All About His Fate03 julho 2024
Where Is Thomas Mitchell Overton Now? Learn All About His Fate03 julho 2024 -
 Blog – William Atterbury, Maryland Immigrant03 julho 2024
Blog – William Atterbury, Maryland Immigrant03 julho 2024 -
 The Project Gutenberg eBook of The Life and Times of the Rev. John Wesley, by Rev. L. Tyerman.03 julho 2024
The Project Gutenberg eBook of The Life and Times of the Rev. John Wesley, by Rev. L. Tyerman.03 julho 2024 -
 William A. Goodman Historic Documents Collection - Manuscript Collection Finding Aids - Dig Memphis - The Digital Archive of the Memphis Public Library & Information Center03 julho 2024
William A. Goodman Historic Documents Collection - Manuscript Collection Finding Aids - Dig Memphis - The Digital Archive of the Memphis Public Library & Information Center03 julho 2024 -
 Staff03 julho 2024
Staff03 julho 2024 -
 Robin Strasser - News - IMDb03 julho 2024
Robin Strasser - News - IMDb03 julho 2024 -
 Angelina College Coach Inducted Into Diboll Wall of Honor03 julho 2024
Angelina College Coach Inducted Into Diboll Wall of Honor03 julho 2024 -
 Our Leadership Team03 julho 2024
Our Leadership Team03 julho 2024 -
Did You Know? Johnson County, TX03 julho 2024
-
 Episode 163: The Hollandsburg Massacre A Storm Rolled In – True Crime Couple – Podcast – Podtail03 julho 2024
Episode 163: The Hollandsburg Massacre A Storm Rolled In – True Crime Couple – Podcast – Podtail03 julho 2024
você pode gostar
-
GitHub - gamejolt/gamejolt: This is the whole frontend for Game Jolt. It powers the site and the client.03 julho 2024
-
 The God of High School Götter Trailer03 julho 2024
The God of High School Götter Trailer03 julho 2024 -
 NEW NARUTO THE MOVIE-Road to Ninja--SasuSaku Scan! by TheUZUMAKIchan on DeviantArt03 julho 2024
NEW NARUTO THE MOVIE-Road to Ninja--SasuSaku Scan! by TheUZUMAKIchan on DeviantArt03 julho 2024 -
 Consulta e Jogos de Tarot Online Grátis e Simpatias. O Tarot03 julho 2024
Consulta e Jogos de Tarot Online Grátis e Simpatias. O Tarot03 julho 2024 -
 Allzone Oficial on X: SPOILERS One Piece 1062 LINK: – Os agentes da CP0 estão vindo para matar Vegapunk, eles possuem ao seu lado um Seraphim com a aparência de Kuma. #ONEPIECE1062 #onepiecespoiler #ONEPIECE03 julho 2024
Allzone Oficial on X: SPOILERS One Piece 1062 LINK: – Os agentes da CP0 estão vindo para matar Vegapunk, eles possuem ao seu lado um Seraphim com a aparência de Kuma. #ONEPIECE1062 #onepiecespoiler #ONEPIECE03 julho 2024 -
![FNF VS Var. (Psych Engine Port) (BETA) [Friday Night Funkin'] [Mods]](https://images.gamebanana.com/img/ss/mods/62c7244cb7555.jpg) FNF VS Var. (Psych Engine Port) (BETA) [Friday Night Funkin'] [Mods]03 julho 2024
FNF VS Var. (Psych Engine Port) (BETA) [Friday Night Funkin'] [Mods]03 julho 2024 -
Read Isekai With Smartphone: Another Fanfiction - Spandam - WebNovel03 julho 2024
-
 Chess Boxing: Meaning, History, Rules & Challenges - Blog03 julho 2024
Chess Boxing: Meaning, History, Rules & Challenges - Blog03 julho 2024 -
 The Eminence in Shadow: Master of Garden】 Drawing a cute girl03 julho 2024
The Eminence in Shadow: Master of Garden】 Drawing a cute girl03 julho 2024 -
 Forza Horizon 5's Next Three Seasonal Updates Detailed03 julho 2024
Forza Horizon 5's Next Three Seasonal Updates Detailed03 julho 2024